Investigating the interplay between DNA damage repair mechanisms, genomic instability and radiotherapy resistance
Primary supervisor: Sarah McClelland, Queen Mary University of London
Secondary supervisor: Kasper Fugger, University College London
Project
Triple-negative breast (TNB) cancer affects over 8,000 people annually in the UK. Mainstay treatments – surgery, chemotherapy and ionizing radiation (IR) – still result in 40% patients suffering disease recurrence, partly due to radiotherapy resistance. Collateral radiotherapy damage in healthy adjacent tissue further restricts effective dosing, despite advances such FLASH irradiation (ultra-fast delivery reducing normal tissue toxicity) or focused proton beams. There is a pressing need to enhance radiotherapy efficacy at lower doses to spare healthy cells and to minimise therapy resistance.
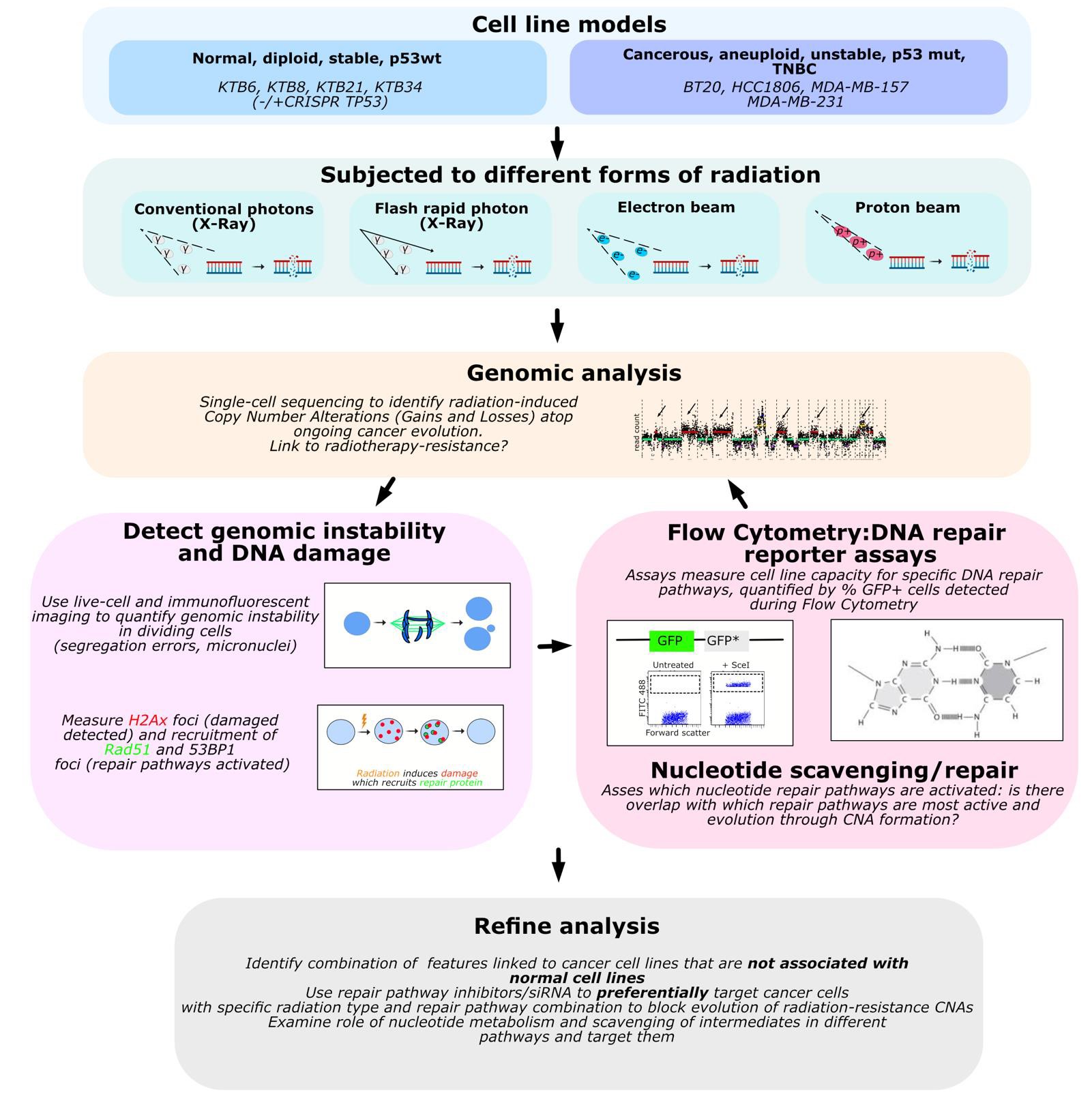
We previously showed that human cells react to genetic insults (such as DNA Replication stress) by forming amplifications and deletions (known as copy number alterations, or CNAs) at vulnerable genomic loci in a reproducible fashion1. Others have demonstrated that unstable tumours, including in breast cancer, display tissue-specific CNA patterns linked to tumour evolution and therapy resistance.2 We hypothesise that radiotherapy itself may induce CNA signatures that promote cancer progression or predict resistance and aim to analyse how different radiotherapy modalities alter the genome of TNB cancer cells.
We will study how radiation induces CNAs, by treating cells with repeated therapy-relevant doses of IR and then using low-pass, whole genome DNA sequencing at the single-cell level to identify de novo genomic alterations. This will tell us: if there is a reproducible pattern of alterations; whether that pattern is related to radiotherapy-resistance; how that contrasts with new CNAs already occurring during ongoing evolution, driven by inherent cancer genomic instability. We will assess the relationship between ongoing instability and inherent resistance (by measuring defects such as chromosome mis-segregation or micronuclei formation in dividing cells).
We will use DNA repair GFP-reporter assays3 (as well as immunofluorescence/live cell imaging of repair proteins recruited to DNA damage foci) to quantify which lesser repair pathways (e.g. single-strand annealing or microhomology-mediated end joining) are preferentially used to repair radiation-induced DNA damage alongside major canonical pathways such as nonhomologous end-joining, and create “repair profiles” for each cell line; we will then test if these can be targeted (using genetic manipulation or chemical inhibitors) to prevent radiation-induced genomic evolution towards resistance-related CNAs. Given the mutagenic nature of IR resulting in generation of modified mutagenic/cytotoxic nucleotides, we will also investigate if nucleotide detoxification pathways are implicated in formation of CNAs and if there is any overlap with the repair profile for each cell line.
We will repeat the above assays with proton beam therapy and FLASH irradiation to assess whether these methods show similar mutagenic CNA formation and therapy resistance compared to conventional IR.
Finally, we will perform parallel experiments in immortalized healthy breast cell lines, to identify therapeutic strategies that preferentially impair cancer cell evolution while sparing normal cells.
Candidate background
This project would suit candidates with a keen interest in a multi-disciplinary project studying cancer therapy resistance through evolution, DNA repair mechanisms, nucleotide metabolism, all through state-of-the-art methods such as single-cell DNA genomics, metabolomics, and high-resolution microscopy.
Potential Research Placements
- tbc, University College London
- tbc, University College London
References
- Shaikh, N. et al. Replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses. Genome Biol. 23, 223 (2022).
- Jamal-Hanjani, M. et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
- DNA Repair Protocols. vol. 920 (Humana Press, Totowa, NJ, 2012).
