Investigating the role of protein synthesis in DNA repair and radiation response to improve cancer radiotherapy
Primary supervisor: Ivana Bjedov, University College London
Secondary supervisor: Susana Godinho, Queen Mary University of London
Project
Radiation remains a key modality for treating cancer. Traditionally, studies of radiation response have focused on DNA doublestrand breaks, in relation to reactive oxygen species and hypoxia. The contribution of damage to other macromolecules and organelles to radiation response has been largely overlooked, as have been some other processes such as protein synthesis.
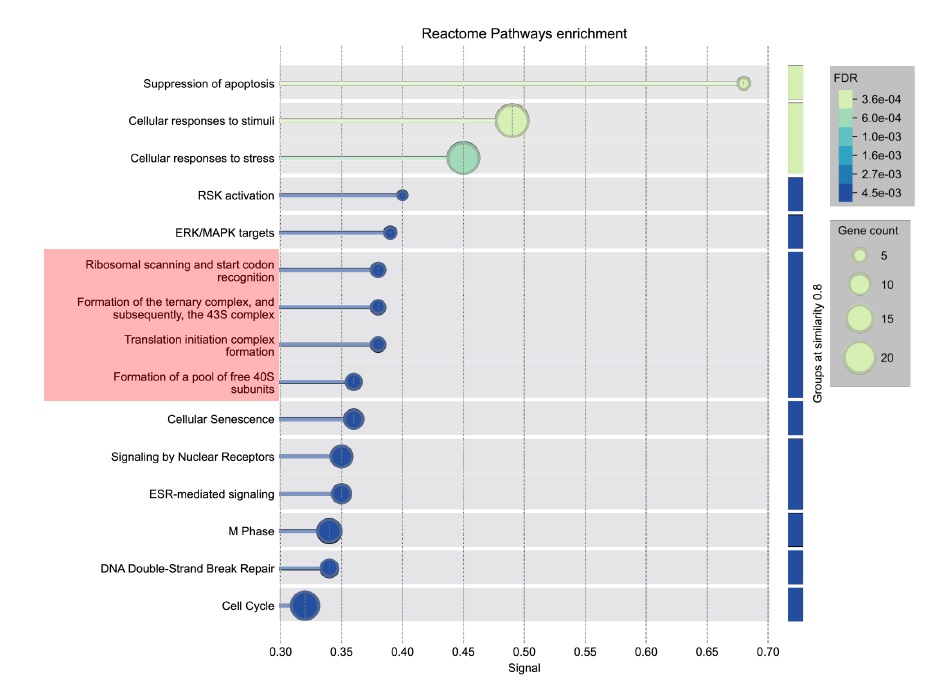
Our re-analysis of a CRISPR/Cas9 screen (Durocher lab) and an siRNA screen (Helleday lab) identified protein synthesis genes as common determinants of radiation survival (Figure 1.). Protein synthesis is frequently reprogrammed in cancer, with changes in nucleolar size, rRNA/tRNA production, and translation initiation and elongation. Onco-ribosomes and ribosomal protein mutations have been reported as oncogenic drivers [1], and 46% of colorectal cancers harbour an oncogenic 18S rRNA variant also found in other tumours [2].
After radiation, protein synthesis is rapidly suppressed to conserve resources, then reprogrammed to support DNA repair. How translation differences in normal and tumour tissue shape radiotherapy response is poorly understood. Notably, eIF5A upregulation in tumours has been linked to radioresistance [3], while the tRNAase SLFN11 promotes ribosome stalling at rare codons and apoptosis upon DNA damage [4].
Together, these findings suggest that components of the translation machinery contribute to oncogenesis and could affect DNA
damage response. We hypothesize that improved understanding of translation changes upon radiation in normal and tumour cells could lead to novel selective targets to enhance radiotherapy.
Aim 1: To use patient-derived assembloids to monitor macromolecular damage and protein synthesis upon radiation. We will build on recent advances in multicomponent 3D co-cultures (patient-derived assembloids) to monitor radiation-induced changes in tumour cells versus matched cancer-associated fibroblasts [5]. Assembloid co-cultures resemble mature crypts and contain both adult stem cells and differentiated cells, resulting from reciprocal interactions between stroma and epithelium. This makes them more physiologically relevant than organoids [5]. Patient-matched assembloids will be obtained from the
Cancer Institute Organoid Facility (Prof. Chris Tape and Petra Vlckova UCL CI collaboration).
To characterise radiation responses in different cell types, we will monitor, at multiple time points, DNA damage (γH2AX), RNA damage (with Dr. Kasper Fugger UCL CI), protein carbonylation (DNP adducts), lipid peroxidation (MDA and 4-HNE adducts), ROS levels (DCFH-DA), and mitochondrial morphology (IF for ATP synthase and PDH) and potential (TMRM). Translation will be measured by puromycin incorporation.
Aim 2: To characterise differential protein synthesis changes in tumour cells and fibroblasts upon radiation. The detailed analyses in Aim 1 will reveal potential differences between tumour cells and fibroblasts and inform selection of the most suitable time point for translational analysis. Ribosome profiling and RNA-seq will be performed separately in tumour and fibroblast cells to identify different proteins and isoforms being translated upon radiation. In collaboration with Prof. Nicky McGranahan (UCL CI), we will apply bioinformatics approaches to integrate these findings with existing datasets and determine which candidates are most strongly associated with poor prognosis and cancer vulnerability.
Aim 3: To validate selected targets pharmacologically and genetically as modulators of radiation response. We will perform pharmacological inhibition and CRISPR/Cas9-mediated targeting of the top 3–5 candidate genes or pathways identified. Epistatic analysis will be used to confirm their role in radiation response, with a focus on selective radiosensitisation of tumour versus normal tissue in assembloids. Fractionated radiotherapy will be employed to better mimic patient treatment regimens.

Candidate background
This project suits candidates with a background in cell biology and interest in cancer and 3D cultures such as organoids and assembloids. Willingness in learning bioinformatic skills is useful and interest in radiation response is required.
Potential Research Placements
- Susana Godinho, Queen Mary University of London
- Nicholas McGranahan, University College London
- Chris Tape, University College London
References
- Howard GC, Tansey WP: Ribosome-directed cancer therapies: the kp of the iceberg? Trends Pharmacol Sci 2025, 46:303-310.
- Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, Spencer Miko SE, Wieden HJ, Landthaler M, Morin GB, Mager DL: Loss of m(1)acp(3)Psi Ribosomal RNA Modificakon Is a Major Feature of Cancer. Cell Rep 2020, 31:107611.
- Zhong Y, Chen X, Wu S, Fang H, Hong L, Shao L, Wang L, Wu J: Deciphering colorectal cancer radioresistance and
immune microrenvironment: unraveling the role of EIF5A through single-cell RNA sequencing and machine learning. Front Immunol 2024, 15:1466226. - Boon NJ, Oliveira RA, Korner PR, Kochavi A, Mertens S, Malka Y, Voogd R, van der Horst SEM, Huismans MA, Smabers LP, et al: DNA damage induces p53-independent apoptosis through ribosome stalling. Science 2024, 384:785-792.
- Lin M, Hartl K, Heuberger J, Beccaceci G, Berger H, Li H, Liu L, Mullerke S, Conrad T, Heymann F, et al: Establishment
of gastrointesknal assembloids to study the interplay between epithelial crypts and their mesenchymal niche. Nat Commun 2023, 14:3025.
